class: title-slide, center, middle <span class="fa-stack fa-4x"> <i class="fa fa-circle fa-stack-2x" style="color: #ffffffcc;"></i> <strong class="fa-stack-1x" style="color:#3b4245;">04</strong> </span> # Data & Workflows ## R Markdown for Pharma ### Alison Hill, Thomas Mock · RStudio #### [https://rmd4pharma.netlify.app//](https://https://rmd4pharma.netlify.app//) --- class: middle, center # <i class="fas fa-cloud"></i> # Go here and log in (free): http://rstd.io/rmd4pharma-cloud --- class: middle, center, inverse <span class="fa-stack fa-4x"> <i class="fa fa-circle fa-stack-2x" style="color: #fff;"></i> <strong class="fa-stack-1x" style="color:#2f5275;">1 </strong> </span> -- # Parameters --- class: left, top ### Knit with Parameters .tiny[ ```` --- title: "Penguins" output: html_document params: species: Adelie --- ```{r setup, include = FALSE} library(tidyverse) smaller <- palmerpenguins::penguins %>% filter(species == params$species, !is.na(body_mass_g)) ``` # Penguins `r nrow(palmerpenguins::penguins) - nrow(smaller)` penguins are classified as `r params$species`. ```{r, echo = FALSE, message = FALSE} smaller %>% ggplot(aes(x = body_mass_g, y = flipper_length_mm)) + geom_point() + geom_smooth(method = "lm") + labs(title = params$species) ``` ```` ] --- 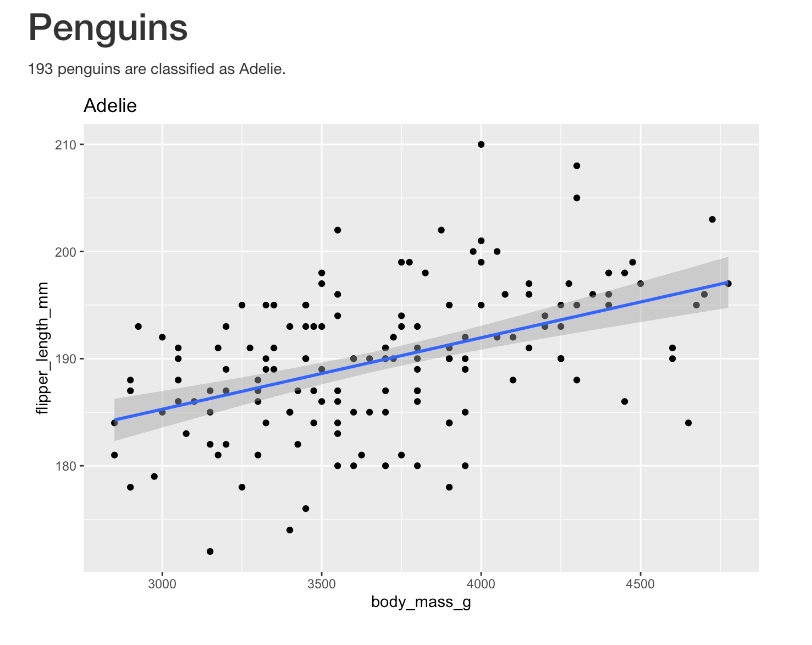 --- class: middle, center, inverse <span class="fa-stack fa-4x"> <i class="fa fa-circle fa-stack-2x" style="color: #fff;"></i> <strong class="fa-stack-1x" style="color:#2f5275;">2 </strong> </span> -- # Command Line --- class: left, top ### `rmarkdown::render()` ```r library(rmarkdown) # needs to be loaded first render("rmarkdown-report.Rmd") ``` -- Now that was easy! But why would you want to use `render` rather than just clicking `knit`? -- How about running a batch of RMDs? + a report + a safety document + a slide deck + or a full website -- Run for each drug of interest *all at once*! --- layout: true <div class="my-footer"><span>https://rstd.io/rmd4pharma-cloud</span></div> --- class: top, left # Render loops -- ```r # the setup unique_species <- unique(palmerpenguins::penguins$species) unique_species #> [1] Adelie Gentoo Chinstrap #> Levels: Adelie Chinstrap Gentoo ``` -- ```r # the loop for(species in unique_species){ rmarkdown::render( "penguin-report.Rmd", params = list(species = species) ) } ``` #### <i class="fas fa-smoking-ban"></i> psst...the wrong way - but what's wrong? -- * What's the name of the output report? --- class: top, left ### We need two things For the `rmarkdown::render()` function to work the way we want: 1. The list of species -> `params` 2. The name of each report -> `output_file` We could do this with a loop, but let's start **purrr**ing... --- class: middle, center > "Of course *someone* has to write **loops**. > It doesn't have to be *you*. > `-` Jenny Bryan --- class: middle, center  --- class: middle, center  --- class: top, left ### Rendering multiple reports ```r # the setup unique_species <- unique(palmerpenguins::penguins$species) # the function penguin_render <- function(species){ rmarkdown::render( "penguin-report.Rmd", output_file = paste0(species, "-report.html"), params = list(species = species) ) } # create the reports purrr::walk(unique_species, penguin_render) ``` --- class: your-turn # Your turn Use this format to create separate site reports for each of the 13 sites in our `mockdata`. Note that there are two main files: - `project/analysis/R/parameterized-rmd.R` -- **Skeleton code** - `project/analysis/reports/site-report.Rmd` -- **Actual report** ```r # the setup unique_species <- unique(palmerpenguins::penguins$species) # the function penguin_render <- function(species){ rmarkdown::render( "penguin-report.Rmd", output_file = paste0(species, "-report.html"), params = list(species = species) ) } # create the reports purrr::walk(unique_species, penguin_render) ``` --- class: middle, center, inverse <span class="fa-stack fa-4x"> <i class="fa fa-circle fa-stack-2x" style="color: #fff;"></i> <strong class="fa-stack-1x" style="color:#2f5275;">3 </strong> </span> -- # Reference external R code --- class: top, left ### Entire .R files `source()` will allow you to read in entire .R files, and is commonly used to *source* external R functions. -- ### Chunks of .R files Adding `## ----` and a name to read chunks by name with `knitr::read_chunk()` ```r ## ---- smaller-penguins smaller <- penguins %>% filter( species == "Adelie", !is.na(body_mass_g) ) ## ---- plot-penguins smaller %>% ggplot(aes(body_mass_g)) + geom_histogram(binwidth = 100) ``` --- class: top, left .pull-left[ ### Parent R Markdown ```` --- title: "Penguins" date: 2020-08-11 output: html_document --- ```{r setup, include = FALSE} library(tidyverse) library(palmerpenguins) ``` ```{r, child=c(“adelie-report.Rmd”)} ``` ```` ] -- .pull-right[ ### Child R Markdown `adelie-report.Rmd` ```` --- output: html_document --- ```{r, echo = FALSE} smaller <- penguins %>% filter(species == "Adelie", !is.na(body_mass_g)) ``` # Penguins `r nrow(penguins) - nrow(smaller)` penguins are classified as `r params$species`. ```{r, echo = FALSE} smaller %>% ggplot(aes(body_mass_g)) + geom_histogram(binwidth = 100) ``` ```` ] --- ## Conditional Child Documents Add specific chunks if conditions met! ```` --- title: "Drug Safety and Response" output: html_document --- ```{r setup, include = FALSE} library(tidyverse) drug_data <- read_csv("drug-data.csv) pass_fail <- summarize(drug_data, mean = mean(toxicity_param)) ``` ```{r, child=if (pass_fail >= 1.5) “safe-drug-addendum.Rmd”} ``` # Dose Response ```{r} ggplot(drug_data, aes(x = drug_cocn, y = drug_response)) + geom_point() + geom_smooth() ``` ```` --- class: left, top ### Links + [Parameterized Reports](https://bookdown.org/yihui/rmarkdown/parameterized-reports.html) + [Parameterized Lesson](https://rmarkdown.rstudio.com/lesson-6.html) + [RMarkdown Cookbook](https://bookdown.org/yihui/rmarkdown-cookbook/) + [RMarkdown Definitive Guide](https://bookdown.org/yihui/rmarkdown/) --- class: inverse, middle, center # Thank you!